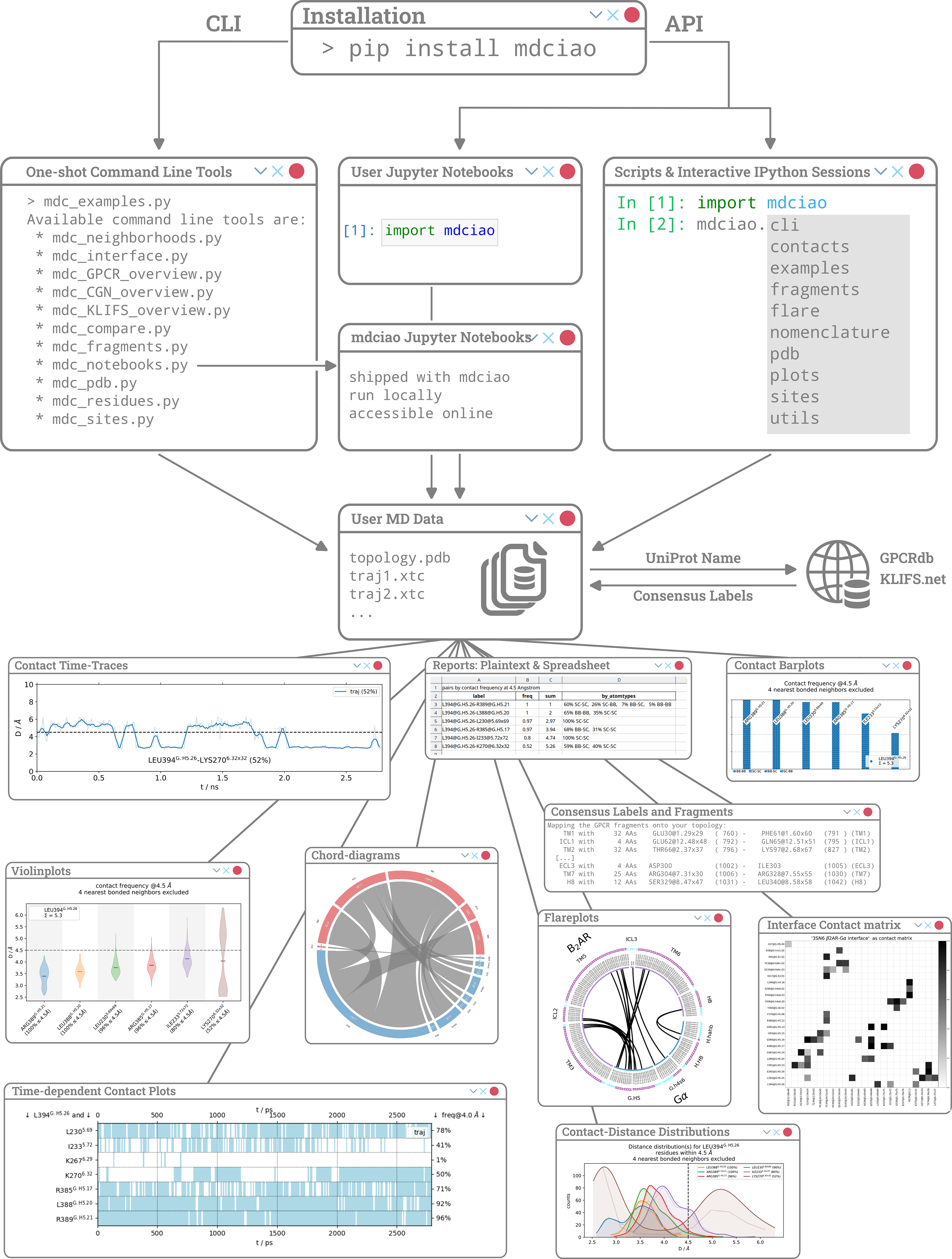
Fig 1: Things to do after installing mdciao.#
DOI#
Abstract#
We present mdciao, an open-source command line tool and Python Application-Programming-Interface (API) for easy, one-shot analysis and representation of molecular dynamics (MD) simulation data. Building upon the widely used concept of residue-residue contact-frequencies, mdciao offers a wide spectrum of further analysis and representations, enriched with available domain specific annotations. The user-friendly interface offers pre-packaged solutions for non-expert users, while keeping customizability for expert ones. Emphasis has been put into automatically producing annotated, production-ready figures and tables. Furthermore, seamless on-the-fly query and inclusion of domain-specific generic residue numbering for GPCRs, GAIN-domains, G-proteins, and kinases is made possible through online lookups. This allows for easy selection and comparison across different systems, regardless of sequence identity, target residues or domains. Finally, the fully documented Python API allows users to include the basic or advanced mdciao functions in their analysis workflows, and provides numerous examples and Jupyter Notebook Tutorials. The source code is published under the GNU Lesser General Public License v3.0 or later and hosted on
https://github.com/gph82/mdciao
, and the documentation, including guides and examples, can be found at
https://www.mdciao.org
